Betraying Mendel to engineer the species - the story of active genetics
With great power, comes great responsibility
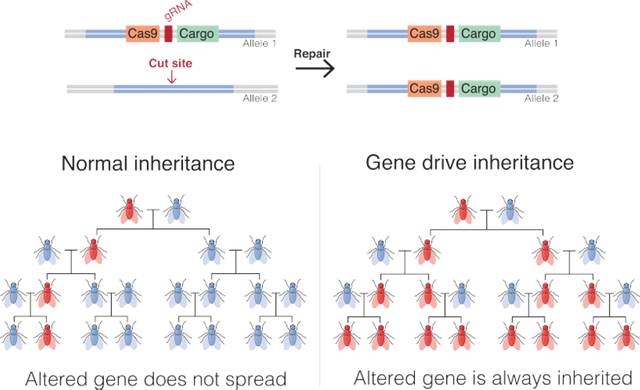
Image by Mariuswalter | CC BY-SA 4.0
The technology I am going to describe today, holds great potential. It can help to eradicate vector borne diseases. Such as, malaria and dengue which spreads by mosquitoes. It can help eradicate invasive species that hinders agriculture. It can help modify not just few organisms, but entire populations and species, to look and behave, the way we desire. Given it holds such power, the onus of using it responsibly also lies on us. Here my job is to make you aware of how it works. Because if society understands technology better, they should be able to make informed choices for their future.
Mendelian genetics for newbies
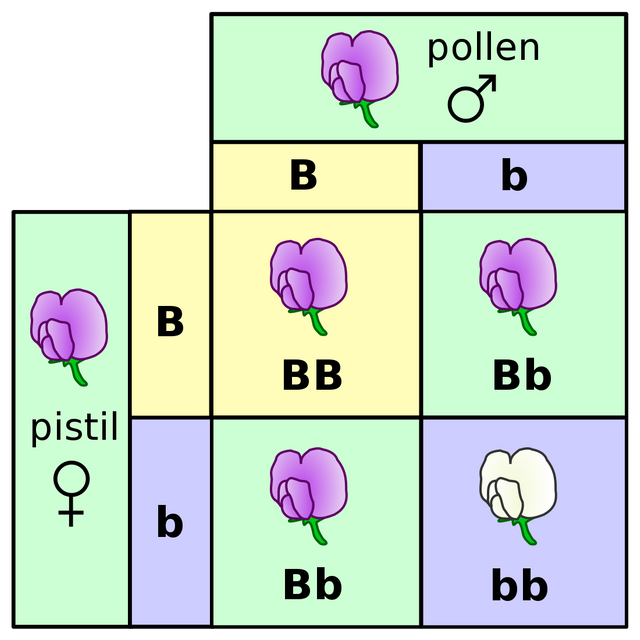
Image by Madprime | CC0 1.0
Every sexually reproducing organism carry 2 copies of genes in their genome, on homologous chromosomes. The two copies of genes can be identical, in which case we will call the animal homozygous for this genetic locus. In other cases, the organism can have similar but not identical sequence of same gene, on their homologous chromosomes. Or you can say that the organism carries two different alleles of the same gene and is hence heterozygous for this genetic locus.
When the sexual gametes form they randomly pick one of the two chromosomes. And hence one of the two alleles from the same chromosome. So if there was a gene G on some chromosome, and g on the other - then 50% of sperms of this organism will carry G and other 50% will carry g. Similarly if the female as well have G and g alleles, then 50% of eggs formed in this female will only have G and 50% of them will have g.
During the mating, there are 4 possible combinations of sperm and egg thar can form - GG,Gg,gG,gg. Therefore, if this male and female pair makes 100 kids in their lifetime 25 of them will be GG, 25 will be gg, and the rest 50 of them will be Gg. If G was a dominant gene which makes the organism green, the 75% of offsprings have GG, gG or Gg will be green. And 25% gg offsprings will be white (see Mendelian inheritance).
Population genetics
.png)
Image by Angelahartsock | CC0 1.0
Say, individuals in a very large population who are randomly mating, and have no selection pressure on their color carry alleles. Say, in this population 70% individuals carry allele, G. Then what is the frequency of different genotypes and phenotypes in the population?
Hardy-Weinberg equation tells us that, in such a situation.
Frequency of:
gg + 2Gg + GG
0.32 + 20.30.7 + 0.72
0.09 + 0.42 + 0.49 = 1 is a population in HW equilibrium. The next generation will now have same allele frequencies. G = 0.7 and g = 0.3. This will continue generation after generation in this population. Unless, the for some reason the gene frequency changes, we will always have 91% green, 9% white organisms in the population - generation after generation. The gene frequency may change because green individuals prefer other green individuals for mating. Or because, either green or white individuals are more likely to die because of some pathogenic disease.
This, would always be the outcome provided every gene on every chromosome behaved in Mendelian manner, and there is no selection pressure. But, what about crossing over and recombination that happens during gamete formation?
Recombining genes

Image by Peter Coxhead | CC0 1.0
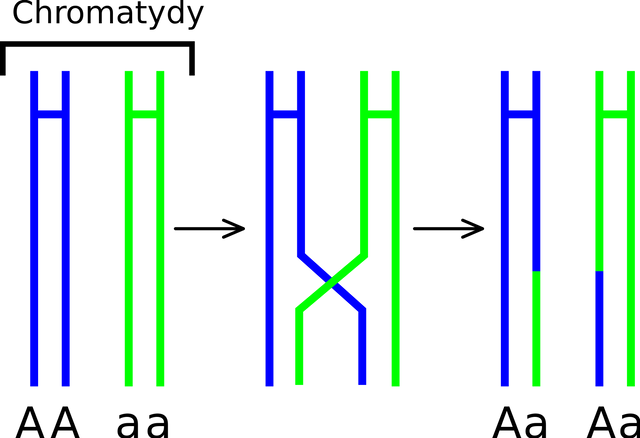
Image by Masur | Public domain.
When the gametes form, they undergo a process called meiosis. In meiosis, the chromosomes segregates and each diploid germ cell forms 4 haploid gametes. Anyhow, during the process of meiosis, the homologous chromosomes come close together. They recognise each other, by their sequence similarity. But this sequence similarity and their closing in, also enables them to exchange DNA and repair each other. In this process called recombination one or more genes on the chromosome cross-over. Which means they bind together by forming bonds between complementary strands. If the mismatch is found, then the DNA repair enzymes copies sequence of one of the gene to other. This creates a possibility that a organism which was heterozygous for some gene, produces gametes of only one of them, as if it was homozygous.
However, for most part cells don't know which gene is the correct one. Which allele will copy onto the other is a random choice. Hence, it a germ cell containing G and g, during a crossover will become homozygous for G or g has equal probability.
So say if chance of crossover at gene locus was 10% then in population there will be individuals which may bias the number of G gametes by 10%. But at the same time there will be individuals who may have biased the number of g containing gametes. Hence, in a large population, and in long run the effect of recombination would cancel out. Yet again maintaining the allele frequencies like they were in the parent generation. However, not always.
Meet the gene drive
What if, during the recombination process g is always biased to replace G? Maybe g even induces the recombination event at this site. Well, if this was to happen, say 90% of the time then - in the next generation the population will have 90% more gametes of g, than expected.
So if frequency of g was 0.3 in the population. Instead of producing. 30% of g gametes were supposed to come from gg and Gg genotypes. But since 90% of Gg's germ cell will change to gg you will be left with 4.2 Gg and 38.8% more gg. Which means that in next generation you will have 49.9 g gametes and only 51.1 G gametes. This will bias the frequency towards g in the population. It will keep increasing in every generation through heterozygotes, until it reaches an equilibrium state; even if being white (gg) was detrimental over being green (GG or Gg). We can simulate different scenarios for fun in another post, if you like.
Anyhow, what we must focus on asking is - do such examples exist in nature? And if yes, then how do we exploit them?
Natural examples of gene drive
There are two major examples of naturally occuring gene drive found in nature.
Transposable elements
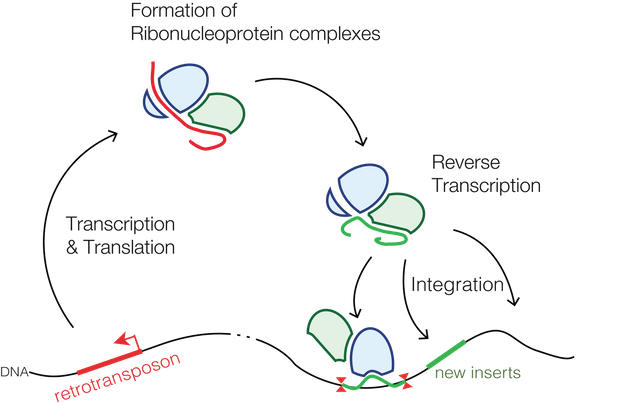
Image by Mariuswalter| CC BY-SA 4.0
Transposons or jumping genes are sequences in the genome, which makes their copies and inserts this copy to a new site in the genome. This increases their frequency in next generation.
Take P element in fruit flies for example. If you inserted a gene of your interest within the P element; and insert that P element into some of the individuals within population, soon the gene will spread throughout the population (Carareto et al., 1997).
Even humans have such jumping genes. Take example of retrotransposons. These are remains of ancient retroviruses (cousins of HIV), that left their genetic material in us. They are well known to move around and expand in our genome - by copying complete or partial gene sequences at new places in our genome. (Janoušek et al., 2016).
Homing endonuclease genes.
.jpeg)
Image by Guido4 | CC BY-SA 4.0
Note that, homologous recombination can also be initiated by damage. That is if DNA has a double stranded break, then it will try to repair it. The repair can happen either by non homologous end joining, or by homology directed repair (HDR). Its is HDR where in order to repair the damage the homologous sequence is used as a template.
Homing endonuclease genes (HEG) produces an enzyme that cuts a specific sequence in the genome as recognized by the enzyme. Once the cut has been made, the double stranded break machinery is deployed to repair it.
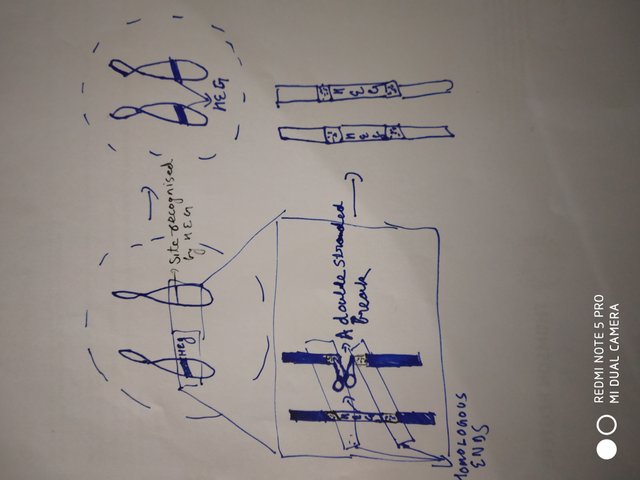
Doodled by @scienceblocks
If the site of cut is spanned by sequence similar to that spanning HEG, then HEG is inserted at the new site. This situation is of course more likely to happen between same locus on two homologous chromosomes (and, it is more frequent during meiosis). This biases the copying of HEG from HEG containing chromosome to non HEG containing chromosome. This ends up increasing the frequency of HEG containing chromosome in the population (Burt et al., 2003). Anyway, what this hints at is that if you control where to make the cut, you can control and bias the recombination of genes.
Active genetics - exploiting the gene drive for human benefits
If nature can do it, so can we. The potential of using gene drive for designing and exterminating wild type insect populations, has been recognized since 1960s (Curtis, 1968). But the biotechnological tools to do it efficiently, only became available in the end of 20th century. It was the time, when restriction enzymes came into picture. I mean imagine if you can attach an HEG gene to a gene of your interest, and span it with sequences that causes recombination at site of your choice (Burt et al., 2003). This should enable you to design artificial gene drives, when you release the lab made populations into the wild. However, you will be limited by constraints that your locus of interest in the genome, should have a DNA sequence that can be recognized by HEG.
You can however overcome this constraint by either mutating the HEG to recognize a DNA sequence of your desire or by engineering TALEN or ZFNs proteins that cut DNA at site you engineered it for. But, if and only if, we had an endonuclease which doesn't have to be designed every time but just programmed to cut at specific site. I guess we know one - they call it Cas9.
CRISPR-Cas9 based gene drives
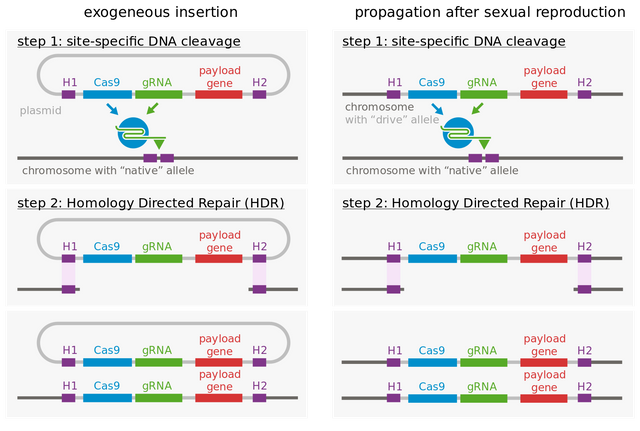
Image by Thomas Julou | CC BY-SA 4.0.
If you are unaware of how CRISPR-Cas9 system works, I recommend you read my previous blog. The crux of the matter is that - you can just design a sequence of guide RNA (gRNA), complementary to site where you want to make a double stranded cut. The Cas9 enzyme bound to gRNA will bind to region where it matches and make a double stranded break. This will start the break repair process. Now if you had your gene if interest fused with Cas9-gRNA genes, bounded by some homologous sequence, the repair machinery will insert this cassette, into the strand it is repairing. And hence, you have created a programmable, and site specific tool to create artificial gene drive. Which brings us to the question of how to use it.
Gene drive user manual - examples
Now that you are aware of tools that can be used to create artificial gene drives, we can visit some examples of its utility.
Manipulating traits
In 2015, Gantz and Bier designed a Cas9-gRNA cassette flanked by sequences homologus to yellow gene found on X chromosomes of Drosophila. The called this gene a mutagenesis chain reaction or MCR. A recessive homozygous female mutant for this gene (in which both copies of gene are mutated) and all male mutants (because they only have one X chromosome), will have yellow colouration in the cuticle of the flies. Under normal scenario a yellow female (homozygous mutant for y, y-), crossed with y+ male, should give all yellow males and no yellow females in 1st generation. In second generation these hetrozygous y+/y- females, with y- males, should yield 25% of yellow females, 25% yellow males, 25% non yellow females and 25% non yellow males. However, for Gantz and Bier who introduced y- mutation by MCR construct, this did not happen. Instead, they got about 49% yellow males, 47 % yellow females. Only the remaining 4% were non yellow or mosiac individuals. If you were to release the MCR containing flies in the wild, in matter of few generations the yellow flies will be all over the pace.
How to eradicate malaria
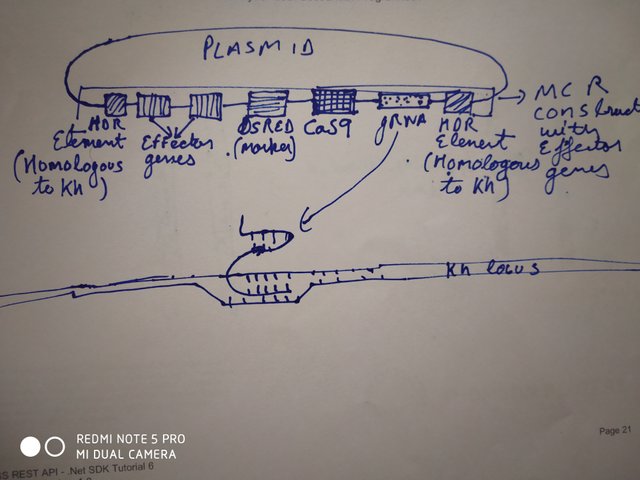
Alright then, if we can spread the MCR construct to modify the color of flies, can we use it for something more productive? How about eradicate malaria? Malaria is spread by female mosquito Anopheles stephensi. There are many proposed ways to spread genes in mosquito populations that would make the females infertile, or change the sex bias to males via meiotic drive. But my favorite one is to use the mutagenic chain reaction construct, carrying the genes that make the mosquitos incapable of carrying the germ, Plasmodium falciparum.
Gantz et al., 2015, designed such a construct. Their construct had 2 effector genes, namely m2A10 and m1C3, both of which disables spores of Plasmodium falciparum to accumulate in saliva of Anopheles stephensi. These genes were fused with gene for red fluorescent marker along with gRNA and Cas9. The gRNA was designed to make Cas9 cut the Kh gene locus in mosquito. Mutation of this gene causes mosquitos to have white eyes. Hence , mosquitos that will have MCR gene will have white eyes. Finally, the entire construct was flanked on both sides by sequence that promotes homology directed repair at Kh locus. Using this construct Gantz and the team were able to make mosquitoes that do not spread malaria and spread this gene through the population at 99% rate.
I can talk about more strategies and examples. But I think you get the point. Using CRISPR-Cas based gene drive we can design the entire wild populations. Be it eradicating diseases, getting rid of pests from our crops, or modifying rodent populations (Grunwald et al, 2019). Whatever, your need be, the gene integration or deletion can be done for entire populations and not just individuals. Though, you are limited by generation time. For instance if you wanted to modify entire human population and gene reached 99% of people in 20 generations, it may take you centuries to achieve your goal.
Another issue, as you might have guessed (and why eradication of malaria has not started in wild), is because of ethical concerns. Just because we can play God with genes, should we do it, is what haunts us. What if we miss something and something goes wrong? I will leave you with that question to wonder about.
This post has been voted on by the SteemSTEM curation team and voting trail in collaboration with @utopian-io and @curie.
If you appreciate the work we are doing then consider voting all three projects for witness by selecting stem.witness, utopian-io and curie!
For additional information please join us on the SteemSTEM discord and to get to know the rest of the community!
This post needs a gene driving a car as top image XD
It did cross my mind. More like a car driving on the gene. 😂 Probably in some other post now.
This makes a good companion article to @agmoore's recent mosquito post I guess, regarding ways to tackle the little peskies.
I've got to read this again:)...after I finish yours.
Hi @scienceblocks!
Your post was upvoted by Utopian.io in cooperation with @steemstem - supporting knowledge, innovation and technological advancement on the Steem Blockchain.
Contribute to Open Source with utopian.io
Learn how to contribute on our website and join the new open source economy.
Want to chat? Join the Utopian Community on Discord https://discord.gg/h52nFrV
Hi @scienceblocks!
Your post was upvoted by @steem-ua, new Steem dApp, using UserAuthority for algorithmic post curation!
Your UA account score is currently 3.423 which ranks you at #7064 across all Steem accounts.
Your rank has not changed in the last three days.
In our last Algorithmic Curation Round, consisting of 208 contributions, your post is ranked at #197.
Evaluation of your UA score:
Feel free to join our @steem-ua Discord server