Strategies for reversing the programming of environment on our genes (Part 2 - CRISPR toys for programming specific genes)
Environment modifies the way genes are expressed. A stressful environment especially during childhood may alter the gene expression profile, in an attempt to make you sturdy and ready for adversities in life. However, this alteration may at times, come with a cost. A cost one may pay with their mental and physical health later in life. But can it be reversed? In this part, we explore the means by which we can specifically control the epigenetic programming (which regulates gene expression in response to environment) for our benefit.

-CRISPR-Cas9's Bohemian Rapsody
Created using following images -
DNA Helix Frame III Vortex by GDJ | CC0 1.0
Target by typographyimages | CC0
Recap
In the previous article, I told you, how epigenetic reprogramming translates the effect of environment to your biology. I discussed two key mechanisms, namely, the modification of histones (the proteins on which DNA is wrapped around and packed), and methylation of DNA at promoter region of genes. We saw that by adding/removing various functional groups to histones such as methyl or acetyl group we can make the packaging of DNA tight or loose. When DNA is wrapped tightly around these proteins the genes are not readily available to other proteins, such as transcription factors (which regulate gene expression) and RNA polymerase (which expresses the gene by copying the genetic code on mRNA). Similarly, methyl group can be added to gene promoters (the region on DNA, where transcription factors bind before they recruit RNA polymerase to transcribe the gene), in response to environment. The methyl group hinders the ability of transcription factors to bind to DNA.
I told you that an adverse stressful environment can alter epigenetic programming, and hence program our gene expression in a way that is harmful to our mental and physical health (also see this blog). Which made many people question that whether this reprogramming can be reversed. Hence, began the exploration. Therefore, in the previous article, I discussed with you some methods that could help us reverse the epigenetic reprogramming; which has been at least shown in animal studies to reverse the effects of adverse environment. The methods we explored included - environmental enrichment, diet and drugs. The crux was that since adverse environment leads to adverse epigenetic reprogramming, positive intervention with relaxation, meditation, vacation, music lessons, etc should have positive effect. And we saw to a degree it does. I also mentioned how components needed for enzymes causing epigenetic modification can be controlled via diet. For instance, controlling methionine amino acid intake in diet and some vitamins can alter the state of DNA methylation.
Nevertheless, in the end of the article we came to a point where we realized that environmental enrichment, diet and drugs are bruteforce methods. For instance, adverse environment may cause methlyation of some genes, while at the same time demethylate others. Some genes like FKBP5 will overexpress, while others like BDNF will be down regulated. Hence, we just can't be sure which diet or drug to play around with. Say, if we were to use drugs to target DNA methylation or histone modification, these drugs would be directed against the proteins such as DNMTs (enzyme that adds methyl group to DNA) and HDAC (enzyme that removes acetyl group from histone). These drugs could either be activators or inhibitors of these enzymes. Nevertheless, activating or inhibiting these proteins, is nothing different from going into a dark room, and randomly pressing switches to turn on the light. In context of cells this may not only be hit in the dark, but can be dangerous. Quite recently, Alexandra Avgustinova showed that, playing around with global histone metylation to cure cancer, may look promising to begin with, but it actually give rise to much more aggressive tumors. Well, this makes our case for not using drug or diet assisted bruteforce rather strong. We need something specific, so we can program the gene expression in a controlled fashion. But, then how do we specifically target the genes?
RNAi can target specific genes.
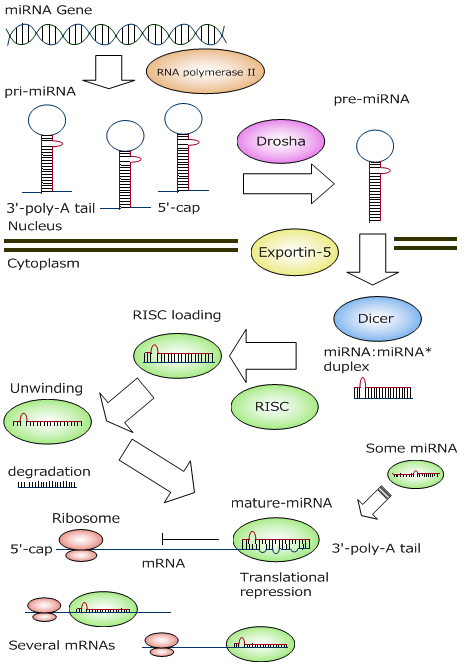
The image shows the natural miRNA that is expressed in our cells. It is produced as a long pre-miRNA and exported out of the nucleus. There it's taken by the group of proteins called the RISC complex. The RISC complex processes it and makes a short mature miRNA. The complex then goes and binds to its target mRNA in its 3' untranslated region and leads to its degradation.
Figure by いいぐさ | CC BY-SA 4.0
Well, one way would be to use RNAi technology. Well, this technique acts after gene has been transcribed into mRNA. But, even mRNA in the cells is regulated by another type of RNA called microRNA or miRNA. If miRNA binds to mRNA it interferes with the translation of mRNA into protein, majorly by targeting the mRNA for degradation. However, we can also externally deliver miRNA and siRNA, and stop a gene from being expressed. Given we can design miRNA and siRNA specific to a gene we can control expression of which gene do we exactly downregulate.
However as you might have guessed, this is only a good tool for genes that get overexpressed and cause trouble, such as FKBP5 for our purposes. This is not an ideal tool of genes such as BDNF, which gets downregulated by adverse life experiences. Moreover, unless I use a modified lentivirus (HIV virus) to deliver this miRNA or siRNA into the genome of the organism, it is going to be a transient downregulation. The actual epigenetic modification which is causing the overexpression of gene of our interest will be unaffected by this method. So, you will keep needing the RNAi dose over and over again to keep this gene in check. Which not to mention will not really be very affordable in the long run. So we need something the hits the core of the matter. And what hits specific gene better than CRISPR?
Let's make toys with CRISPR-Cas9
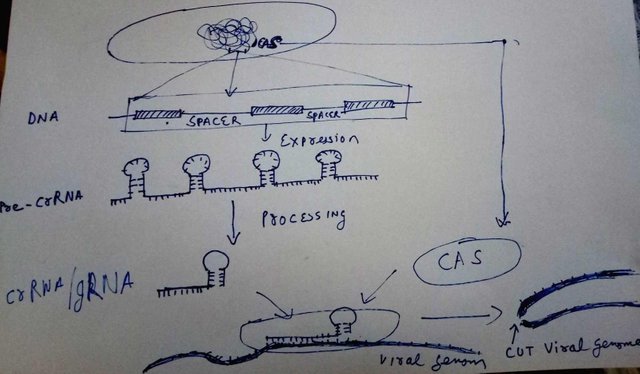
Doodled by @scienceblocks and also used in my previous blog
CRISPR technology has been very successful in targeting specific genes. In an earlier post on CRISPR-Cas, I talked about how it is the adaptive immunity in bacteria. And, more importantly how we can use it to edit/delete/add genes. The essence of the matter is that bacteria stores tiny pieces of genome of viruses that infected it in, a special region in DNA called CRISPR. The region contains repeated palandromic sequences with a spacer region after each repeat. It is in this spacer region where the sequence of viral DNA is stored. So if the virus happens to infect this bacteria again the RNA made by this region will bind to it.
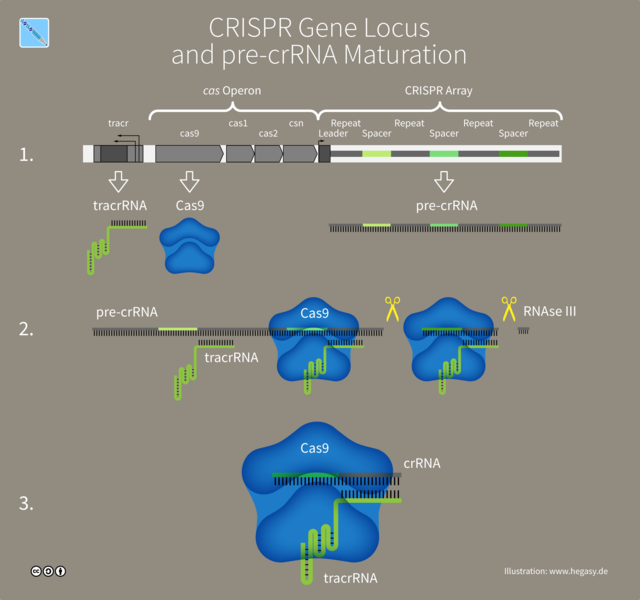
Figure by Guido4 | CC BY-SA 4.0
The whole CRISPR region in bacteria is expressed as a single pre-crRNA. Another piece of RNA called transactivating RNA or tracrRNA , binds to an enzyme, Cas9 and activates it. The activated Cas9, then binds to pre-crRNA and cuts it into individual crRNA with help of tracrRNA. Each crRNA have it's individual region containing the piece from viral genomic sequence, and another from the reapeats that forms the stem loop. The loaded and activated Cas9 enzymes then goes for a hunt and if any DNA sequence matches the viral sequences it just cuts it into two peices.
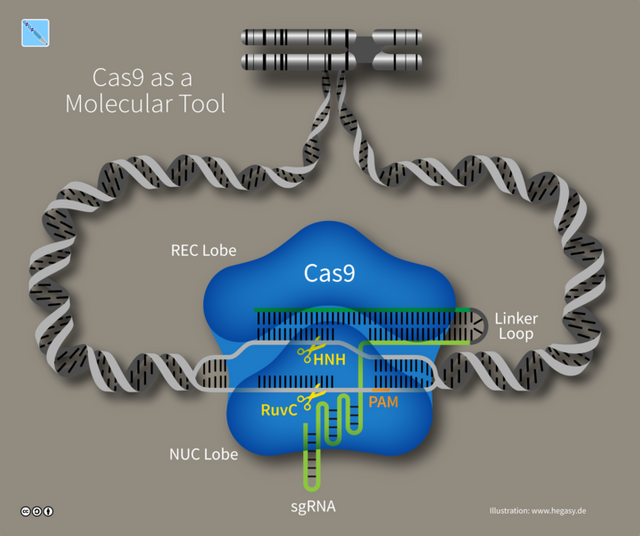
Figure by Guido4 | CC BY-SA 4.0
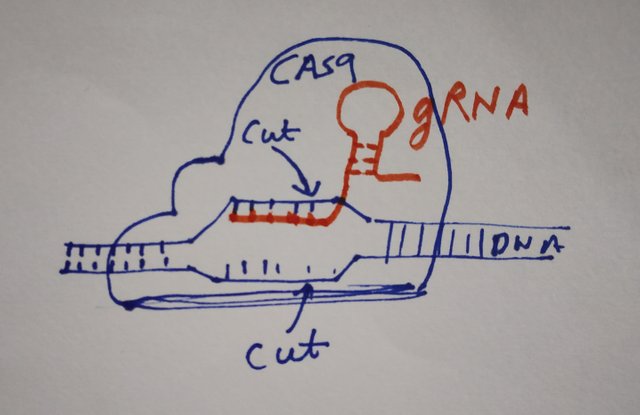
All you need to do to make a system for editing genome is design a special kind of RNA, called the guide RNA(gRNA) or short guide RNA (sgRNA) which will specifically bind to your gene of interest. And then an enzyme called Cas9, which specifically binds to this gRNA will cut the DNA, right where your gRNA will bind. If you are lost and wondering what's the difference between crRNA-tracrRNA vs gRNA, them gRNA is just a fused RNA that contains both of them in a single piece. Well this just makes our life easy in terms of using the technology.
Using this method you can make cuts in the genome at specific sites, and even insert new DNA peices between these cuts of you like. However, can we use the specificity of gRNA and modify the Cas enzyme in a way that instead of cutting the DNA, it rather modifies the epigenome at specific sites? Turns out you can. But, how! you ask. Well to understand that we need to revisit CRISPR technology in a bit more detail and understand how Cas9 enzyme works. So what say, are you ready to take a dip in details? Come on, it will be fun.
Anatomy of Cas9
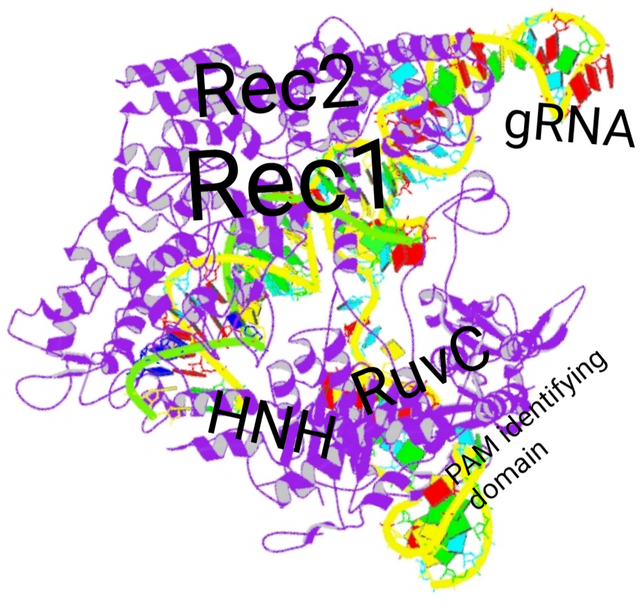
Adapted from PDB structure database PDB ID:4OO8 , as described in publication by Nishimasu et al., 2014 | License information for using PDB data
While some proteins might have a single domain and perform single function. But, many proteins are like these complicated nano-machines, where different parts or domains of the protein have specific function. A lot like your smartphone. You smartphone have different parts - camera, screen, fingerprint reader etc, bound into single entity. Cas9 too is a nano-machine with different domains. It can be divided into two functional lobes - the recognition lobe or Rec and a nuclease lobe or Nuc. The Rec lobe is responsible for binding to gRNA and activating the Cas9. While Nuc lobe is responsible for cutting the DNA which binds to gRNA by complementary base pairing. The REC has REC1 and REC2 domains. And, the NUC lobe has HNH and RuvC domain. The HNH domain cuts the strand of DNA that is bound to gRNA, while the RuvC domain cleaves the strand that isn't. Together they make a double stranded break in the DNA (see Jinek et al., 2014). The two lobes are connected via bridge linkers, the details of which don't matter for us now. And, there is yet another domain in Cas9 called PAM interacting domain, we back to this later.

Pictures taken by @scienceblocks
For now, all I want you to think of Cas9 is that it is a nano-machine, like some fancy scissors. In fact, if you think of scissors, its handle would be the recognition lobe, in which you can insert a part of your guide RNA, which will be your fingers. Once the fingers are in, the scissor is active. The blades of the scissors are like nuclease domain. You guide it to where you want make the cut in some thread, and snip!
Re-engineering Cas9
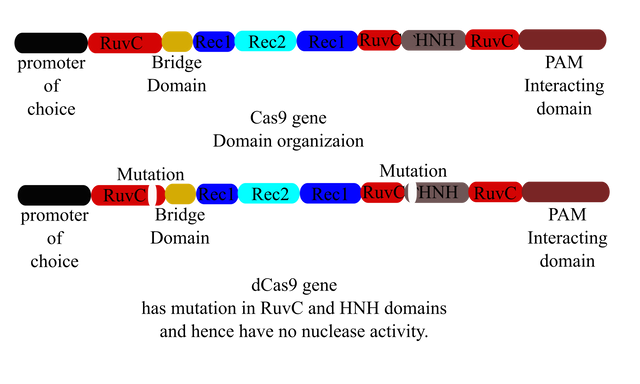
Illustrated by @scienceblocks
Cas9 gene architecture drawn from information provided in Cavanagh & Garrity, “CRISPR Mechanism”, CRISPR/Cas9, Tufts University, 2014.
Now, if I wanted to have a paint brush which I can indeed hold like scissors, I can just tape the damn blades together, and stick a brush at tip. So now I have a tool which I can hold the same way with my hand as guide, but won't cut the thread. It would sure as hell paint the thread.
I won't go into structural details, but you can think of epigenome modifying enzyme as paint brushes. The paint here would be the methyl or acetyl or whatever chemical group. And the canvas is either the DNA or histone proteins. All this brush is missing is that handle that can precisely guide it to the gene where I want to put the paint. But, what if I just fuse the recognition domain of Cas9 with DNMT or HDAC. Sounds like cool toy.
I can take care of the fact that it won't cut my genes by mutating or even deleting the nuclease domains. One such mutant of Cas9, which binds to the gRNA and helps Cas9 to be recruited at target gene but doesn't make cuts is called dCas9 or catalytically dead Cas9. Well, we can use that. All we need to do is create a gene that expresses dCas9-epigenome modifying enzyme as a fusion protein.
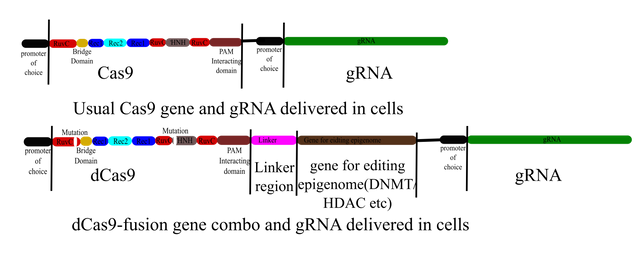
When editing genome Cas9 gene with a gRNA targeting the desired gene is delivered into the cell. When editing epigenome a dCas9-epigenome editing enzyme fusion gene can be delivered to cells also with the guide RNA to target specific site for epigenome editing.
Illustrated by @scienceblocks
VS
dCas9-DNMT
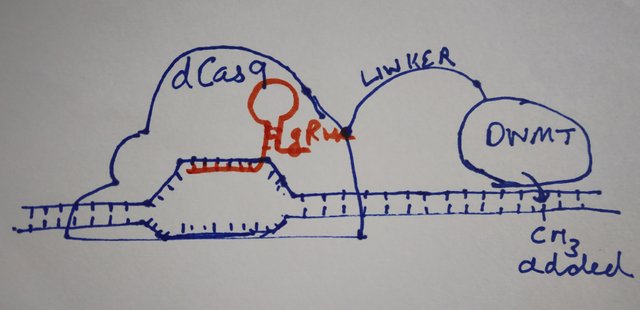
The dCas9 part of the fusion protein, binds to the location on DNA as specified by gRNA. The DNMT3A part of the protein in the meantime methylates the nearby cytosine residues in CpG sequences.
doodled by @scienceblocks
For example, Liu et al., in 2016, created few such toys. They fused DNMT3a (enzyme that methylates the DNA at recruited site) with dCas9 and showed that using gRNA guided against specific locus they can methylate the gene of their choice. They were successful in demonstrating this not only in cells but also in brain of live animals. They also created another toy where they fused Tet (an enzyme which demethylate DNA) with dCas9 and successfully demethylated the targeted gene. In another study Lei et al., fused dCas9 with an efficient methyltranferase from bacteria - MQ1 DNA methyltranferase, and used it to methylate the eukaryotic genes in targeted manner. So here are examples of a few toys you can use to specifically edit epigenome. There are in fact even more fancier things that people have made with Cas9, but I will keep that for some other day. In the meantime aren't you wondering if anyone has also edited histone modifications using CRISPR?
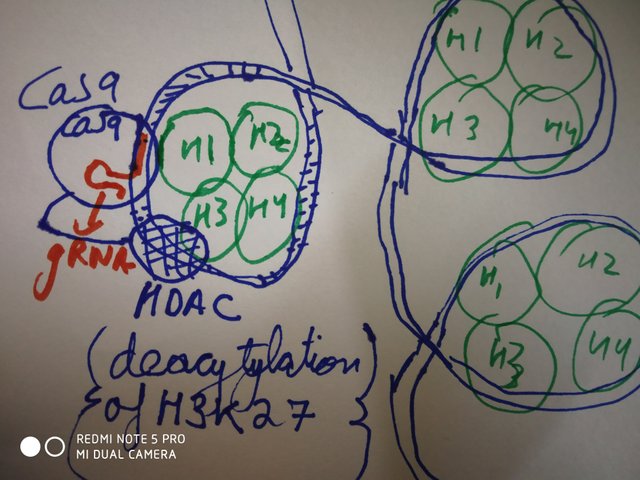
doodled by @scienceblocks
Turns out, people have been trying fusion of Cas9 with different histone modifying enzymes, in this regard. For instance Kwon et al., in 2017, fused an engineered version of HDAC3 (enzyme which removes acetylation from lysine at position 27 in histone H3, and hence represses gene expression), with dCas9. Though, they found it little difficult to find exact site to design the target gRNA against. Well because chromatin state would interfere with gRNA binding. But nonetheless, with little permutation and combination they found the right spot on DNA that would cause deacetylation at site of their choice. On the other hand, Hilton et al., in 2015, fused dCas9 with p300, which has opposite effect of HDAC3. That is p300 adds an acetyl group to lysine number 27 in histone H3 and activates the genes. In yet another study performed in 2015, by Thakore et al., , they fused dCas9 with KRAB (an enzyme that add 3 methyl groups to 9th lysine in histone H3, and represses gene expression). Doing so, they successfully targeted the gene eenhamcer or their choice, and modified its pakaging. As a result they repressed the target gene.
But, this is just the beginning. Given all different kind of histone modification that we need to build toys for, we have just scratched the tip of the iceberg. Have a look at this table of histone modification and their functions, if you are interested in knowing the kind of molecular toys that need to be made.
Getting rid of PAM limitation.
For those who know a bit more about CRISPR, must be familiar with the limitations of making gRNA, imposed by PAM site. For those who don't know the Cas9 enzyme not only finds a target on DNA complementary to sequence encoded in gRNA, but this target region in DNA is should also have a neighbouring PAM site. PAM site is a 5'-NGG-3' sequence of DNA where N can be any nucleotide. This is a self safety mechanism of bacteria that uses CRISPR-Cas system, so that Cas9 doesn't cut its own DNA. Well bacteria that has CRISPR system in it, lacks PAM sites in its genomic DNA. However, this also limits us to what can we target. Because everytime you will need to make a gRNA, you will need to make sure that it has a neighbouring PAM site. Well, not anymore. This month Chatterjee et al.,, discovered a variant of Cas9 in Streptococcus Canis, which they are calling scCas9. scCas9, unlike regular cas9 requires only 5'-NNG-3' sequence nearby. Well, it gives much more genomic coverage and freedom to design gRNA to desired target genes.
Maybe 50 years down the line you not just be sequencing your genes, but a doctor may also write you tests to sequence your epigenome - maybe RNA-seq, and then bisulphite sequencing and CHIP-Seq for histone modification. Then a bioinformatics guy, will analyze your results and recommend which genes need to be methylated of demethylated. Or, which histone modification should the doctor make. The doctor would fix a date with you, order the toolkit with specific gRNAs. Well, let's just say if I can get my hands on shares of a pharma and healthcare company investing in CRISPR, I am so buying it. I understand that some parts in this post might have been bit intense for those not from biology background. Though I did try to simplify it as much as I could. However, if you did find anything confusing or that you did not understand, please ask me in comments. With this note, I'll leave you here.
About steemstem
But before I go, I would like to mention about the steemstem platform. Well, if you love reading and writing interesting science articles @steemstem is the community on steem that support authors and content creators in STEM field. If you wish to support steemstem do see the links below.
You can vote for steemstem witness here -
Quick link for voting for the SteemSTEM Witness(@stem.witness)
Delegation links for @steemstem
(quick delegation links: 50SP | 100SP | 500SP | 1000SP |5000SP | 10000SP).
Delegating to @steemstem gives ROI of 65% of the curation rewards.
References
Journey from adaptive immunity in bacteria(CRISPR) to saving Man's best friend(Dog).
Cavanagh & Garrity, “CRISPR Mechanism”, CRISPR/Cas9, Tufts University, 2014.
Lei et al., 2017. Targeted DNA methylation in vivousing an engineered dCas9-MQ1 fusion protein
Chatterjee et al., 2018. Minimal PAM specificity of a highly similar SpCas9 ortholog
Signing off
@scienceblocks
I don't have a biology background, just some very basic neurobiology courses in university. I must say that there were many unknown "words" for me here. However, this metaphor simply saved my life :P
And then it got even better!!!
It sounds like a very promising field. If I get it right, this is the bright future of medicine! It is an entirely new approach - eliminating what is working unproperly by reprogramming it. Now we just treat symptoms most of the time. I just hope it happens sooner than 50 years. I would love to see it :D
P.S. Btw, loved your drawings. Too complicated for me to understand, but add a very personal touch :D
Indeed, even I would like to see it happen before 50 years. And it may. In the previous post I described that how they fixed dogs with a muscular degeneration disease using this technology and even removed HIV from animals. It will become mainstream sooner or later. The human trials have just began. I wish they succeed.
An yeah, I realized that there was indeed a lot of jargon in this post. I was trying to capture the details. But someday I will sure write a simplified version of it. Because what's the point of communicating science if I can't get it to reach every person. I am happy to get this feedback esp on the metaphors. I know now how to simplify it further. Thank you so much.
I am really happy that you found my comment helpful!
I am looking forward to your next article!
See you!
Yes, yes, yes :D Can't wait to read it. My little one just woke up, so I will have to leave it for tomorrow. But just wanted to leave you an enthusiastic note :P
😀 Thank you. Now I am definitely enthusiastic to know your response after you read it.
Hi @scienceblocks!
Your post was upvoted by Utopian.io in cooperation with @steemstem - supporting knowledge, innovation and technological advancement on the Steem Blockchain.
Contribute to Open Source with utopian.io
Learn how to contribute on our website and join the new open source economy.
Want to chat? Join the Utopian Community on Discord https://discord.gg/h52nFrV
This post has been voted on by the SteemSTEM curation team and voting trail in collaboration with @utopian-io and @curie.
If you appreciate the work we are doing then consider voting all three projects for witness by selecting stem.witness, utopian-io and curie!
For additional information please join us on the SteemSTEM discord and to get to know the rest of the community!
Hi @scienceblocks!
Your post was upvoted by @steem-ua, new Steem dApp, using UserAuthority for algorithmic post curation!
Your UA account score is currently 3.400 which ranks you at #7031 across all Steem accounts.
Your rank has dropped 84 places in the last three days (old rank 6947).
In our last Algorithmic Curation Round, consisting of 339 contributions, your post is ranked at #255.
Evaluation of your UA score:
Feel free to join our @steem-ua Discord server
Congratulations,
you just received a 12.32% upvote from @steemhq - Community Bot!
Wanna join and receive free upvotes yourself?
Vote for
steemhq.witnesson Steemit or directly on SteemConnect and join the Community Witness.This service was brought to you by SteemHQ.com