Determination of Three Dimensional Structures of Proteins
There are several methods used to determine the structure of proteins. These are X-ray crystallography, NMR spectroscopy and electron microscopy. Each method has advantages and disadvantages. Scientists use a lot of information to build the final in each of these methods. This is the X-ray diffraction pattern for X-ray crystallography. For NMR spectroscopy, this information is the local conformation and the distance between other nearby atoms. For the electron microscope, the entire shape of the molecule must be visualized.
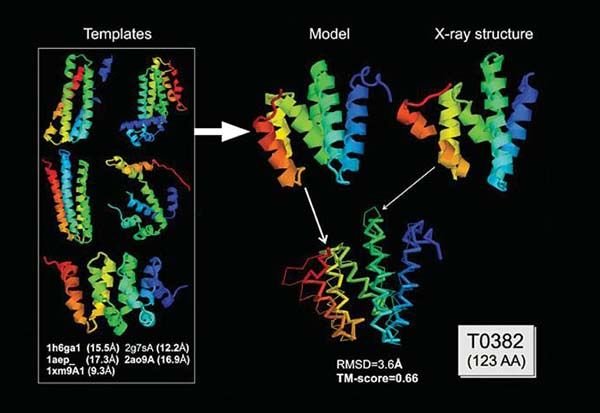
This experimental knowledge is not enough to draw a model from scratch in most cases. Additional information about the structure of the molecule should be added. For example, the sequence of a protein amino acid sequence is the geometry of the atoms in the protein. This information allows scientists to construct both experimental data and the molecule consistent with the expected conformations and geometries.
X-Ray Crystallography
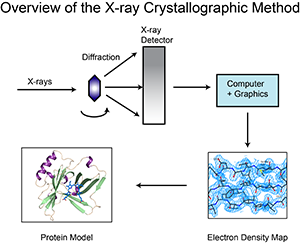
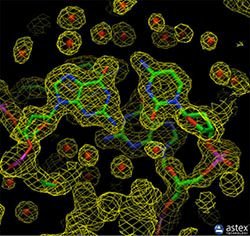
Most of the structures were determined by X-ray crystallography. The protein is purified, crystallized and then extensively exposed to X-rays for this method. The map of the electron density obtained is followed by two types of data for the crystal structures in the archive to determine the position of each atom. These are the coordinate and structure factor files. Coordinate files contain atomic locations for the latest model, data files, structure factors of structure design. An image of the electron density map can be generated using tools such as the Astex viewer via a link in the structure factor file.
X-ray crystallography can provide very detailed atomic information showing each atom, ligand, atomic details, inhibitors, ions, and other molecules incorporated in the crystal in a protein or nucleic acid. However, the crystallization process is difficult and may limit the types of proteins that can be studied with this method. For example, X-ray crystallography is an excellent method for determining the structure of rigid proteins that form regular crystals. It is much more difficult to study flexible proteins with this method, because crystallography is based on the alignment of many molecules in the same direction. Flexible portions of the protein may not appear in the chromatographic electron density maps because the electron density can spread over a wide area. Biological molecular crystals are selective. Some form excellent, well-regulated crystals, while others form weak crystals. The accuracy of the determined atomic structure depends on the quality of these crystals. Two important components of the correctness of a crystallographic structure are the resolution that measures the amount of detail that can be seen in the experimental dataset and the R-value (in the structure factor file) that measures how well the atomic model is supported by experimental data.
NMR Spectroscopy
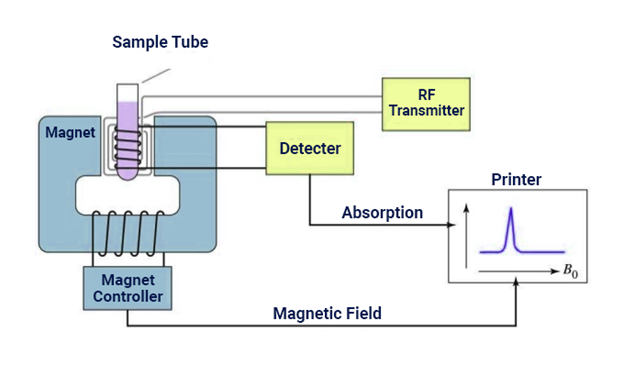
NMR spectroscopy can be used to determine structures of proteins. The purified protein is placed in a strong magnetic field and is investigated using radio waves. A distinct set of observed resonances can be analyzed to give a list of atomic nuclei close to each other and to characterize the local conformation of the atoms connected together. This restriction list is then used to create a protein model that shows the location of each atom. This technique is currently confined to small or medium sized proteins because large proteins have problems with overlapping peaks in the NMR spectra.
The most important advantage of NMR spectroscopy is that it provides information about the protein dissolved in a crystal, as opposed to those bound to a crystal or bound to a microscope grid and thus NMR spectroscopy is a leading method for studying atomic structures of flexible proteins. A typical NMR structure involves a number of protein structures, all of which are consistent with the observed list of experimental limitations. The structures in this community are very similar in regions with strong limitations, and very different in less restricted regions of the chain. Probably with less restriction these areas are flexible fragments of the molecule and therefore do not give a strong signal in the experiment.
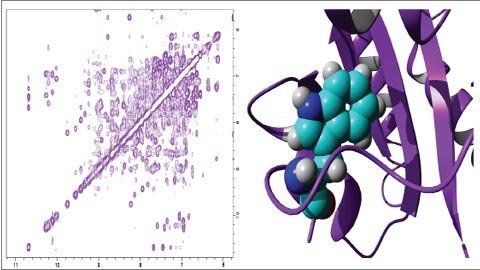
There are two types of coordinate input archives in the archive. Each structure is represented as a separate model, including the full collection of structural determinants in the first one. The second input type is a mean structure that is reduced to a minimum. These files show the average properties of the molecule based on different observations in the community. There is also a list of constraints determined by NMR. These include hydrogen bonds and disulfide bonds, the spacing between adjacent hydrogen atoms, and the localization of the chain and the restriction on its stereochemistry.
Electron Microscopy
Electron microscopy is used to determine large macromolecular complex structures. An electron beam is used to direct the image of the molecule. If the proteins are coaxialized to form small crystals or packaged symmetrically on a membrane, electron diffraction can be used to create a 3D density map using methods similar to X-ray diffraction. If the molecule is very symmetrical as in virus capsids, many separate images can be obtained that provide many different images. These views are then aligned to extract 3D information and averaged. Electron tomography on the other hand, obtains multiple views by rotating a single sample and taking several electron micrographs. These views are then used to give 3D information.
Electron diffraction produces atomic level data in a few system but typically electron micrographic experiments do not allow the investigator to see each atom. Electron micrograph studies often combine the information from X-ray crystallography or NMR spectroscopy to order atomic details. Atomic structures are placed on the electron density map to form a model of the complex. This has proven to be very useful for multimolecular structures such as complexes of ribosomes, tRNA and protein factors, and muscle actinosin structures.
Image Sources: 1, 2, 3, 4, 5, 6

img credz: pixabay.com
Nice, you got a 9.0% @minnowbooster upgoat, thanks to @kedi
Want a boost? Minnowbooster's got your back!
The @OriginalWorks bot has determined this post by @kedi to be original material and upvoted(1.5%) it!
To call @OriginalWorks, simply reply to any post with @originalworks or !originalworks in your message!
They are getting the resolution of Cryo EM down really far these days, the size limitations of yesteryear are not as large of a concern. A few groups are using fusion constructs of a protein of interest with a known oligomeric protein and are using the symmetry to define more accurate structures as well. Theres a reason why this stuff won a nobel prize this year. :)
There's some truly amazing studies being done in Japan by Nakamura where they use EM to watch molecules crystallize. https://www.nature.com/nmat/journal/v11/n10/full/nmat3408.html
@originalworks
Nice post. Have you ever done protein NMR? I've never understood how you could get meaningful structural data from something with so many protons.
Really Its Very Helpfull for Our Life Its Good Information.
Excellent comments & observations !!
really nice post. resteem this post. thanks
Congratulations @kedi, this post is the second most rewarded post (based on pending payouts) in the last 12 hours written by a User account holder (accounts that hold between 0.1 and 1.0 Mega Vests). The total number of posts by User account holders during this period was 2024 and the total pending payments to posts in this category was $1820.13. To see the full list of highest paid posts across all accounts categories, click here.
If you do not wish to receive these messages in future, please reply stop to this comment.