(Re-)Create Life in a Lab - Part III

DNA is an amazing molecule on many levels. It is the support of genetic information in all known organisms, from the simplest bacteria to the most complex organism and it contains all the instructions for a cell to function. We are now so used to the famous double-helix structure that we often forget it was discovered only 65 years ago and is still holding many secrets.
Outside of its obvious aesthetic aspect, the DNA structure also exhibits unique chemical properties allowing the stable conservation of the genetic information, its reading and decoding and its replication to transmit this information throughout cell generations. But DNA molecules are also prone to modifications: their structure could be modified to allow an additional layer of regulation to gene expression and they can mutate to allow the evolution of the genetic information they carry. This equilibrium between stability and plasticity has allowed the evolution of all the organisms we know from a common ancestor. But is the genetic code as we know it the only possible one?
Replication, transcription and translation
DNA molecules are chains of blocks called nucleotides. Each nucleotide is composed of a 5-carbons sugar (deoxyribose), a nitrogenous base and a phosphate group linking the nucleotide to the rest of the chain. Four different nitrogenous bases are naturally found in DNA: Adenine, Thymine, Guanine and Cytosine (respectively abbreviated A, T, G and C), they are the “letters” composing the genetic information. These bases are complementary two by two: each A on one DNA strand is associated with a T on the other strand and each G is associated with a C.
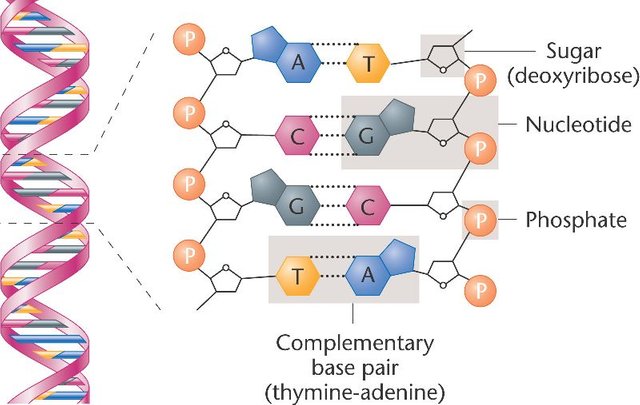
This complementarity allows the reading and replication of the DNA molecule: by opening the DNA double-helix, proteins called DNA polymerases can read each letter on one DNA strand and synthesize the second strand by pairing each base with its complementary one. If you want to learn more about the discovery of the replication mechanism, I encourage you to read this great article by @alexander.alexis.
The base complementarity also allows the decoding of the genetic code by the cell to synthesize proteins that will perform different cellular functions. This decoding starts with the transcription mechanism that creates a molecular “negative picture” of the DNA molecule under the form of a single-stranded RNA molecule (also composed of nucleotides slightly different for those composing DNA). The RNA molecule is then translated into a protein by a large molecular complex called ribosome that reads RNA bases 3 by 3 thanks to adapter molecules called tRNAs. Each of these base triplet (or codons) encode for a specific amino acid composing the protein and the amino acid corresponding to each codon is carried by a specific tRNA. Amazingly, this correspondence between base triplets and amino acids, forming what we call the genetic code is found in every organism on Earth.
As we saw, the complementarity of bases two by two is the essential characteristics allowing the replication and decoding of the genetic information. Would it thus be possible to add new letters to the code? This is the question Floyd E. Romesberg and his team at the Beckman Center for Chemical Sciences (La Jolla, California) tried to answer.
The Universal Genetic Code / Image Source
Creating “Alien” DNA
The team of Pr. Romesberg created in 2014 the first semi-synthetic organism with a genome composed of more than four letters. A new pair of potential complementary bases not found in living organisms, called d5SICS and dNaM, was already identified in 2012 and these bases were shown to be replicated just like natural ones by a DNA polymerase in vitro. The replication machinery indeed does not really care about the chemical composition of the bases it inserts in the newly synthesized DNA strand. The incorporated base simply needs to be complementary to the one that is read and their interaction must conserve the size and shape of a normal DNA double helix. Pr. Romesberg thus added these modified bases to a synthetic DNA molecule (plasmid) and inserted it into bacteria. The team then provided the unnatural bases as independent building blocks to the cells so that they can replicate the plasmid. The transformed bacteria were shown to grow normally while replicating the modified plasmid, becoming the first organism to replicate a 6-letters DNA molecule.
However, these new letters were inserted in an exogenous piece of DNA not coding any essential information and the cells were not able to be decode them to form proteins. Just like you would be able to copy a text written in a foreign language without understanding it, the cells were simply matching each base to its cognate one but did not have the right adapter molecules (tRNA) to translate them into amino acids. This was until 2017, when Pr. Romesberg’s team repeated the experiment but this time provided the cells with tRNAs recognizing the unnatural bases and carrying modified amino acids. This was enough to make the cells translate the new letters into unique proteins containing blocks never observed before in nature.

What “Alien” bacteria tell us
This breakthrough is not only a beautiful proof that we can create simple organisms with unique characteristics, but it also opens amazing new possibilities in synthetic biology.
First, organisms with a unique DNA composition normally not found in nature are not able to synthetize their own nucleotides. This means that they are not capable of proliferating in absence of a source of unnatural nucleotides, which is reassuring in a case where one of these bacteria would escape the lab where it was created. Pr. Romesberg also hopes that bacteria with an extended genetic code encoding a larger set of amino acids will be used to produce original therapeutic molecules. As Romesberg says:
“If you’re given more letters, you can invent new words, you can find new ways to use those words and you can probably tell more interesting stories.”
Lastly, this shows that Life can take other forms than what we find on Earth. We can imagine cells on other planets using other molecules than DNA as a support of genetic information.
However, these experiments were all performed with exogenous DNA that was not inserted in the bacterial genome. The central position of DNA in most cellular mechanisms makes it difficult to imagine an organism able to function correctly with a genome containing unnatural letters. Furthermore, some scientists think that our cells evolved to work with a code of only four letters because increasing the number of different bases would also increase the possibilities of non-specific interaction between two bases from different pairs, which could result in an important imprecision in replication, transcription and translation mechanisms.
While we may not see synthetic monsters roaming the streets any time soon, these experiments question our definition of Life and help us to understand the essential components of living organism on Earth and what an alien cell could possibly look like.
Sources and further read
- The publication of the first replication of DNA with modified bases in vitro
- The first replication of DNA with modified bases in bacteria by the team of Pr. Romesberg
- The publication of the semi-synthetic bacteria able to produce proteins incorporating unnatural amino acids
- A press article asking interesting questions on artificial bases and the evolution of the genetic code
Check out my previous articles
- My introduction
- [CRISPR 101] What is CRISPR?
- [CRISPR 101] DNA breaks, Gene Expression and DNA repair
- Do Lobsters hold the secret to immortality?
- (Re-)Create Life in a Lab - Part I
- (Re-)Create Life in a Lab - Part II
- Are Dogs friendly because of a mental disease?
thanks for that interesting post! im excited which synthetic organism will be cloned next!
Glad you liked it :)
This is such an awesome study! Imagine the implications of expanding the coding power of DNA by a factor of 9! The first thing I thought when you introduced 'alien DNA' was that it would be useless for an animal without the relevent transciption and translation mechanisms to process it to protein, but the fact the team managed to create these too is phenomenal!
When you say
Is this because they introduced DNA in plasmids, and not into the central genome of the bacteria?
I guess the next steps are moving that "alien DNA" into the bacterias central genome. From there someone will have to figure out how to make all this work in Eukaryotes with their increasingly complex and fickle DNA machinery!
I don't envy the team that tries to solve that puzzle!
Thanks for such an entertaining read!
Yes! Just the fact that replication and transcription of unnatural bases can occur without any modification to polymerases is already amazing, especially considering that the bases they use interact mostly through hydrophobic interactions and not hydrogen bongs like normal Watson-Crick pairs. Apparently, just using a normal tRNA with modified anti-codon was enough (they use the seryl-tRNA synthetase from E.Coli which apparently does not require codon/anti-codon interaction so tRNA loading was not an issue).
The DNA template was indeed a plasmid, I also wonder what would happen if you try the same in genomic DNA. I would instinctively say that it would be a mess but if replication and transcription work fine then maybe it would not be that bad in bacteria if you only modify ORFs and not promoters or regulatory elements (eukaryotes are another story though, I'm not sure this would ever be possible).
Glad you liked it, I enjoyed writing it :)
Thanks for elaborating and yeah, loved it :) Keep it coming :)
just WOW.
Introducing alien DNA is one thing and amazing itself, but making it to code for the insertion of non-proteinogenic amino acids takes it to a whole different level!
Yes i remembered reading the first paper in 2014 where they just show replication of the plasmid and it was already impressive. I found out about the second one while writing the article and it blew my mind! I think now the most difficult part is specific tRNA loading of different unnatural amino acids in the same cell, but that's definitely exciting just as a proof of concept.