Targeted Protein Labeling Through Amber Codon Suppression Technology
Hello everyone, I would like to take a brief moment to thank all of you who continue to follow me despite my prolonged periods of absence. I spend a lot of my time working behind the scenes for the @steemstem project and this leaves me with very minimal free moments to construct a post. So I truly appreciate those of you who stick around and visit what I have to share with you. You are awesome and I do not take you for granted.

Todays Blog: Targeted Protein Labeling Through Amber Codon Suppression Technology
Today I will discuss with you all an article published in the journal Nature Scientific Reports titled "Site-Specific Labelling of Multidomain Proteins by Amber Codon Suppression."
In Brief
This article discusses the implementation of amber codon suppression technology, which is a method in which the tRNA encoding for the amber codon (UAG) is replaced with one which allows for the incorporation of a non-natural amino acid. The purpose for incorporating non-natural amino acids into proteins here were the reactive chemical groups they contained. These non-natural amino acids could then react with and allow for the direct attachment of a fluorescent molecule specifically to only one location in the protein of interest.
This enables a whole host of protein biochemistry possibilities to better understand the dynamic nature of how a protein moves as it performs its function in a cell (whether that function be enzymatic (ie. performing a chemical reaction) or structural (ie. serving as a scaffold upon which other proteins can sit)). There are a host of standard biochemical techniques which are commonly done to label proteins for study in this way, however these methods have caveats that limit their utility for certain proteins (eg. those with many cysteines). Making the method the authors devised here, intriguing and potentially very useful for proteins which have been difficult to study to date with more conventional labeling strategies.
If this seems like it might be something of interest to you, by all means, read on and enjoy. If this seems a bit too overly technical, I understand that as well. In which case you likely learned a bit from the summary (yes, people do make glowing proteins, and they do really use that to understand how proteins move!) thank you for checking out the post, and I hope to see you next time I publish!

Traditional Protein Labeling Strategies
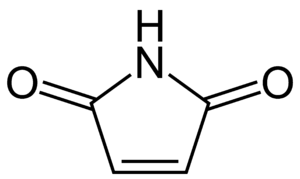
There are a variety of common laboratory methods for putting a fluorescent tag on a protein. The simplest of which is the maleimide labeling strategy. A maliemide group is a chemical functional unit which undergoes a conjugation reaction with a thiol group (-SH). Conveniently proteins have these groups as they are a part of the amino acid Cysteine and are available for reaction when the cysteine is in the reduced state. Many proteins completely lack cysteines, and thus through molecular biology and site directed mutagenesis technology researchers are able to site selectively add one where ever they want. This allows for the researcher to then add a fluorescent molecule which possesses a reactive maleimide group, and presto, you have a protein with a fluorescent molecule attached in a time frame of 30 minutes in a biologically relevant pH (7.4) buffered solution.
However many proteins have cysteines, and many of those have more than one, making this labeling strategy difficult as you will be placing fluorescent molecules all over the place on the proteins surface. Thus there are other labeling strategies such as use of fluorescent molecules with succinimidyl ester groups which permit conjucation with free amines (like the N-terminus of the protein). However this labeling strategy can also allow for labeling of lysines (it has some nice amines to react with, and depending on your buffer conditions they will be very tasty to the succinimidyl ester, you likely don't want that) and labeling at the N-terminus is not always useful.
So big proteins with many cysteines are often difficult to fluorescently label in such a way so as to permit highly quantitative biochemical analysis of their movement dynamics (need to have labels in just one specific spot, otherwise all the movements mix together and you end up being unable to see anything at all).
Enter The Strategy of Today's Article's Authors
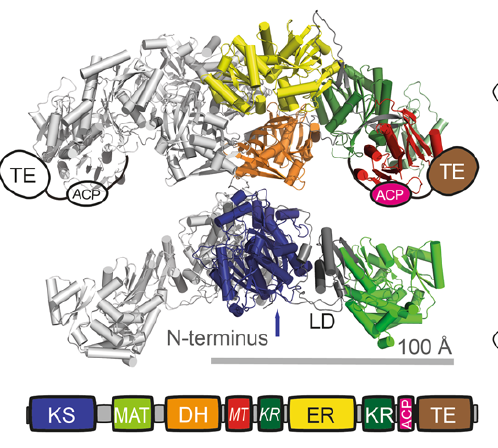
As I mentioned above, they were using amber codon supression technology to add a variety of non-natural amino acids to a big protein which is difficult to study because it has a lot of cystines. The protein they were studying is called fatty acid synthase I or FAS (Seen To The Right) a multi enzyme protein which does what its name implies, synthesizes fatty acids (specifically palmatate).
FAS is a really large protein existing as a dimer (there are two copies of the protein together) and weighing about 540,000 grams/mole enzyme. It possesses multiple active sites which each perform separate enzymatic reactions in a sequential order passing its substrate from one site to the next. This means there are a lot of motions going on which are just begging to be understood (some of which have been observed using atomic force microscopy [2], but this technology lacks the resolution to really understand the proteins workings, for that we need to be able to see single molecules and for that we need fluorescent tags!).
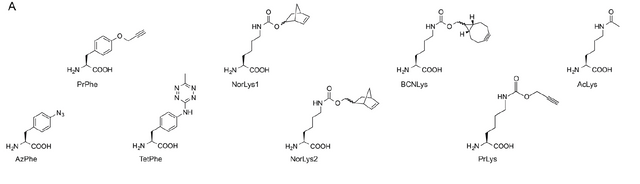
This protein however is not amenable to traditional cysteine labeling strategies (it has far too many native cysteines) and amine labeling wont put fluorophores where we need them to understand anything! Thus the authors turned to site specific labeling through introduction of non native amino acids. They stuck with 8 non natural amino acids (image to the left) which all allowed for the possibility of the attachment of a fluorescent molecule through click-chemistry.
Can The Protein Be Expressed When Non Natural Amino Acids Are Included?
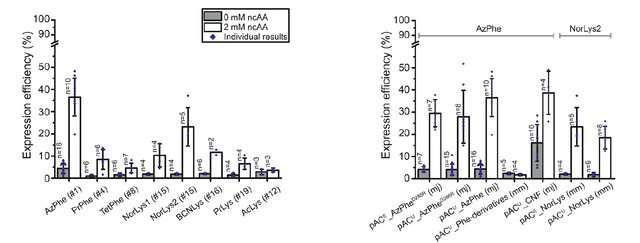
Relative Expression Levels of ACP Protein Constructs Containing The Non Natural Amino Acids: Reproduced From [1] Figure 2B
Prior to building this system into the complicated FAS protein, they began with a more simple system called acyl carrier protein or ACP. It is a small protein which carries a 4-phosphopanthene molecule and is also involved in the synthesis of fatty acids (like the FAS protein they want to study). ACP protein constructs co-expressing a green fluorescent protein were devised containing the non-natural amino acids through the previously mentioned and described amber codon suppression methodology, and protein expression (based on the fluorescence intensity of the green fluorescent protein) was attempted and compared to that of an wild type enzyme (one that doesn't have a non-natural amino acid incorporated).
What they see (image above) is that when the non-natural amino acid is included in the media of the E. coli they expressed the protein in (white bars, the grey bars lack the amino acid and represent the background) that protein expression levels are actually pretty bad for all amino acids except AzPhe and NorLys1 amino acid constructs (left plot). They tried this in a few different expression systems (right plot, including with a a different tRNA from M. jannaschii with a mutation D286R (marked mj) and also with a different expression plasmid pACu). Nothing really further increased expression efficiency much beyond ~40% of WT levels.
Does It Matter Where They Put The Non Natural Amino Acid In The Protein?
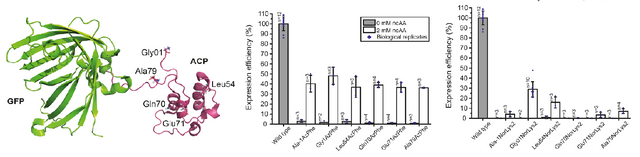
Reproduced From [1] Figure 3A and 3C
They made constructs expressing these non-natural amino acids in a variety of locations and they found that expression levels were relatively consistent for the AzPhe (middle graph) but not so for the NorLys2 (right graph). You can see the locations where they put the amino-acids illustrated on the protein structure (right image) with each spot labeled with a name that is then included in the titles on the x-axis of the two bar graphs.
Bigger Protein Expression
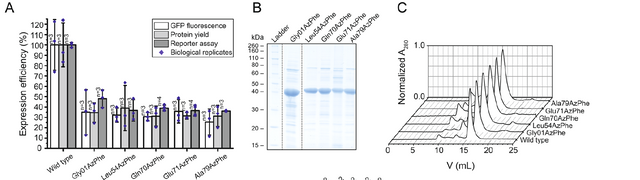
Reproduced From [1] Figure 4A, 4B and 4C
The researchers upped the size of their cultures and really tried to express and purify these proteins, above we are looking at some examples of the AzPhe containng ACP protein. The right plot is the same GFP fluorescence that we have seen in the past. The middle plot is an acrylamide gel, which allows us to separate proteins based upon their size as they migrate through the gel, you can see the blob positioned around the spot marked 40 KDa is the ACP protein, and that the amount that is produced varries a bit with the location where they put the non-natural amino acid. Finally on the right we are looking at a chromatography purification method, where the protein is pushed through small sugar beads which also separate things by their size (size exclusion chromatography) you get a good picture of the size of the proteins that are coming out, based on the amount of volume of buffer it took before they eluted from the column. We can measure what is coming out as proteins have a natural light absorbance characteristic at 280 nm, so monitoring for that absorbance as things elute creates the image that you are seeing in the plots. We see that all of the protein constructs express with a major population that is very similar in size (thats good).
ACP Works and AzPhe is a Pretty Good Choice for a Labelable Non-natural Amino Acid, Will This Work For The Big FAS Protein?
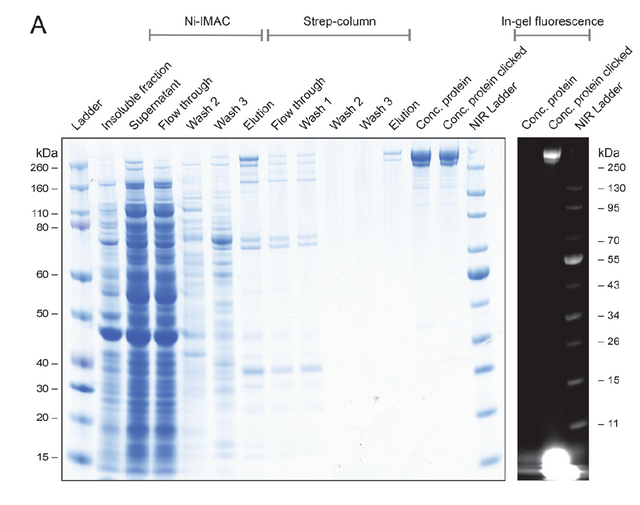
Reproduced From [1] Figure 5A and 5B
Above we are looking again at an acrylamide gel of some expression and purification of a modified version of the FAS protein which contains an AzPhe at position Gly2113. They did a lot of purification work and are showing it on this gel, but the important things for you are the last two lanes labeled Conc Protein (concentrated protein) and Conc Protein Clicked. The Conc Protein lane shows the monomeric form of this protein in all its massive 270,000 gram/mole glory (there is a marker of 260 KDa by it, which is just a bit lower than the big protein blob). The next lane is the same protein that has been reacted by a fluorophore (BCN-649, a dye that absorbs 665 nm light and will emit 676 nm light) that can do the click chemistry reaction with the non-natural amino acid added. In the gel to the left, those two blobs look the same size which is normal. However what if they look at those gels when blasting them with the 665 nm light and measuring the amount of 676 nm light emitted? Well then you look to the right part, where the glowing is only observed in the Conc Protein Clicked lane, indicating that they successfully expressed the non-natural amino acid AzPhe and labeled this massive protein where they wanted it.
Nice Work!
Concluding Remarks
The next steps in this research are to continue testing the properties of this labeled construct, and if they are good, to eventually begin using this labeled construct to better understand the movements of the massive Fatty Acid Synthase enzyme complex. This work marks the very first time that this protein has been selectively labeled, and provides evidence that this methodology could work very well for the study of other large, previously unstudyable proteins.

Other Cited Materials
Images
1.) The horizontal rule used throughout this post is provided for free use by syntaxxx.com
2.) Maleimide Group Reproduced From Wikipedia
Text Sources
1.) https://www.nature.com/articles/s41598-018-33115-5
2.) https://pubs.acs.org/doi/10.1021/acsnano.7b04216

Call To Action
Do you enjoy this sort of post? Do you want more people on STEEM who can do a way better job at this than I can? Well consider giving a vote to the @steemstem project's witness @stem.witness to further enable the team to support the Steem blockchain, and better fund development of their own Steem frontend (dApp) https://steemstem.io
Greetings @justtryme90 !Very interesting and promising this work, I understand that the mechanism of action is similar to that of tumor markers? and on the other hand, would the function of the protein be affected?
Excellent question!! The function of the protein can very much be affected, this is a possibility in any covalent modification of a protein, and even more so by the addition of a non natural amino acid. However one can do a variety of standard biochemical assays to probe enzyme function and if things do not appear significantly perturbed (greater than a 5% deviation from wild type rates) then we would likely suspect that the labeling is not causing a significant difference. However we would not actually know! That's one of the drawbacks to studying things this way. We have no better way to see these movements really, so it's a drawback that we have to mitigate as best we can!
You must continue with the investigation, suddenly you will find new answers and serendipities....
Well someone must at least :D
I have heard of staining (don't know if I'm using the right word) of a particular protein to enable for tracking. This new technique looks innovative. Kudos to people working on the field. I wish them good luck into unravelling more medical mystery and hopefully, one day, arrive on the cure for cancer.
Staining isn't the right word, tracking is always done by some form of labeling. Depending on what you are tracking and where it is usually with a fluorescent molecule as I described above. If you are looking at cellular protein flux (ie production and breakdown of protein/or a substrate over time) then sometimes isotopic labeling can be employed (giving a cell an amino acid enriched with a heavy version of one of it's atoms like carbon 13 or nitrogen 15, or a heavy substrate molecule similarly labeled) and following things by mass spectrometry.
I have a feeling I may be wrong there, but thank you for clearing that up.
You weren't wrong, just not the term for what you were thinking of. :) Perhaps, slightly less right is the better way to think about it!
A little less right sounds much better :)
Posted using Partiko Android
As a biologist who's never paid much attention to protein labelling techniques I found this very interesting. The techniques I'm more familiar with involve attaching a gigantic tag to the protein (which I have always been surprised how it doesn't seem to affect the protein function that much), so attaching a much smaller change is obviously a more attractive solution. The only thing I wasn't sure about was how they make it specific to the protein of interest, apparently what they do is remove the amber codon (which is already not very common) from the entire rest of genome!
This also ignites fever dreams of expanding our genetic code beyond the traditional 20 for no reason in particular other than sounding awesome. "Oh, honey, tell me again how big your genetic code is?" "36, baby. 36."
You are talking about tagging by expressing with like a GFP. That can and does affect protein function very very much in many cases! Steric hindrance alone of the presence of the massive fluorescent protein tag can block protein protein binding sites, make conformational changes difficult and slow substrate binding and turnover rates.
Every change we make to a protein can have far reaching effects, some of which are actually somewhat small, and when looking at a cell, may not be seen until weeks have passed, when all of the sudden the cells get 'sick' due to an imballance in a pathway.
:)
Unfortunately this doesn't get around that caveat! Detailed stufy of a proteins function is necessary to prove you havent perturbed it too much with your label.
Yeah, I know, but it's still surprising that as best we can tell this or that animal/cell is able to live with no problems and the protein carry out its function even though it's dragging behind a big, fat fluorescent tag!
In many cases yeah it is interesting. :)
Finally something worth reading!
I feel ashamed for doing only spin-labelling (*put the label. Done!)
:) thanks. I was at one time a somewhat popular science blogger on here :D
If only I were a good writer!
XD
As one who uses labeled proteins (albeit they are mostly "just" antibodies), but never really spent more than a brief thought on how they are labeled, I have to say this was interesting to read.
There is actually quite a bit of neat chemistry going on even in standard protein labeling techniques! Actually there is quite a bit of neat chemistry going on everywhere, we just don't think about it much of the rime.
Thanks for checking the post out Sco. I went much more technical in my explanation this time, though I still summarized a lot and omitted big chunks of info from the paper for the sake of brevity and telling a story.
I really enjoyed reading this. I’m hoping to see how this turns out. I’m hoping more experiments are carried out and this method is fine tuned and ultimately, widely accepted. It appears to have little to no downsides.
It is a fairly simple approach, and should enable better mechanistic biochemistry on large proteins. Yeah, was a really nice paper.
I must confess, this post is truly technical. Biochemists are trying oh. But the summary was okay though..
So glowing proteins help scientists know how protein move :)
I intended it to be technical :D
Yes, we can use the glowing proteins, and very powerful microscopes capable of looking at one single protein, to monitor how these little molecular machines move and shake.
It was too technical for my comprehension @justtryme90 but I did pick up the bit about making protein's glow, and that also you have some interesting voters (dna-polymerase!?) :)
You get those same voters every time my account votes for you :)
They are my little DNA replication buddies.