The bugs and debugging of DNA - a quick refresher for DNA damage repair
The DNA stores all the information we need to make life possible. It is a code made of four letters A, T, G and C. The sequence of these four letters , contains information on which protein will be encoded. Proteins are these molecular machines that does things for us - from replicating the DNA, to deriving out energy from food and also they make structures that makes cells and tissues looks the way they do. If the sequence in the DNA is misspelled, it will distort the structure of the protein, many times in a way that will wreck havoc on the organism. Hence the information in DNA is sacred, and every effort should be made to preserve it. In this post I will try to explain the threats to this information in DNA and how cells take care of it.
Why am I writing a post?
Why did I decide to do a post about a topic that you may find easily in The molecular biology of cell by Bruce Albert? Well, in the previous post I talked about DNA damage and repair in context of ageing. However, because I was so focused on ageing, I ended up using a lot of jargon and completely ignored the fact that repair mechanisms needs their own explanation. In this article, I will try to explain as simply as I can - the mechanisms of DNA repair. However, before I can explain you how DNA is repaired I should take some time to explain the components of DNA to you. Well, you should have some perspective of what you are fixing. No?
The nuts and bolts of DNA
Illustrated by @scienceblocks
nucleotide image by Calibuon at English Wikibooks | Public domain
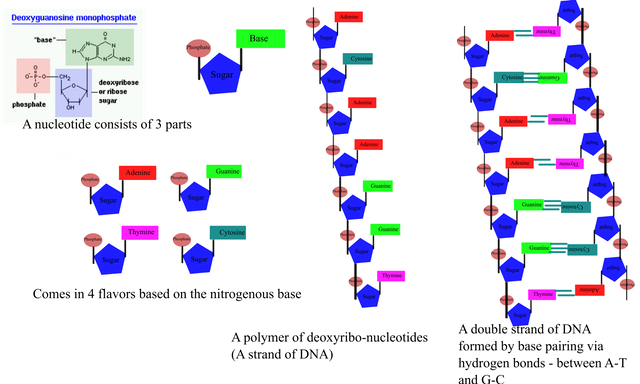
The nucleotides
DNA or deoxyribonucleic acid a polymer of nucleotides. Nucleotides consists of 3 parts - A sugar (deoxyribose for DNA), a phosphate group and a nitrogenous base. Nucleotides that make up the DNA comes in 4 flavours, based on the base that the sugar and phosphate is attached to - adenine, thymine, guanine and cytosine. These nucleotides bond together, forming a chain of up to milliions of nucleotides - the DNA strand.
Every nucleotide has a soulmate
Each DNA stand has a complementary pair - for every A in one strand there is a T, and for every G, there is a C. A-T and G-C pair via hydrogen bonds, forming a DNA double helix. Having the two complementary strands gives the benefit is having an extra copy of information. So in case something happen to one strand, all you need to do is look at the other strand, use it as template and add complementary nucleotides to the damaged strand.
The fuck ups and fixes of DNA
It's like having a office where all the information is managed books/files/scrolls. So each DNA strand is is long sheet of paper (made by sugar and phosphate), and let right page has exact complementary information to the left page. And every time you have to make a new office (cell division), all you need to do is make a copy of these documents and multiply your offices. And this is where the error first starts to appear, for the first time.

Did get your soulmate base? Use the mismatch repair pathway
Let's say you were in prep for making a new office. All you needed was copy of documents. You call your secretary ask him/her to look at the documents and type them out on new sheet of paper. In case, of DNA the name of the secretary is DNA polymerase, but we can call her pol. But no one is perfect, pol makes errors. Sometimes instead of C for a G on template strand she mistakenly puts a T. But, Pol is not that dumb, either. She proofreads it before editors comes and check for mistakes. But, once in a while she misses that mistake. Hence, you need someone who is dedicated to the task of finding a mismatch in two complementary strands.
In our cells the task is assigned to set of proteins involved in mismatch repair. When DNA replication is going on, these protein machines constantly scans the DNA for any mismatched pairs. If A-C instead of A-T is found, or G-T instead of G-C is found, they cut and chew a few nucleotides from the new strand carrying the error, and another Polymerase is assigned to fill the gap. Another, enzyme ligase then comes and seals the strand that fills the gap with the strand in place. Here is a good animation, explaining the same.
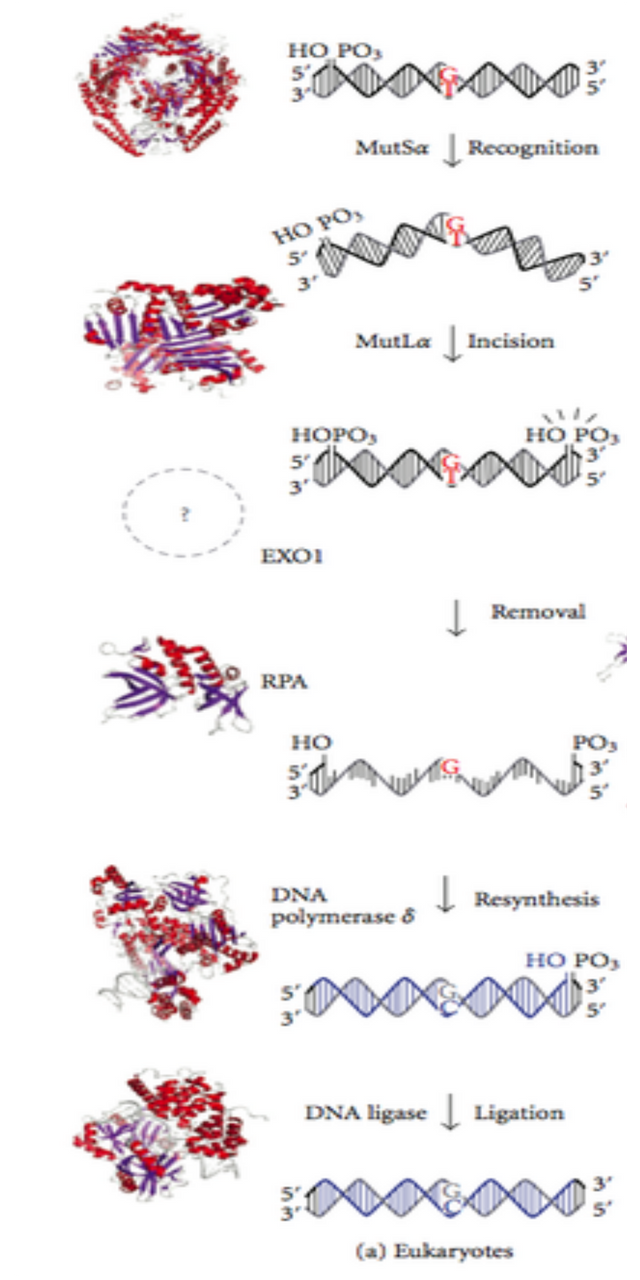
Image adapted from Kenji Fukui | [ CC BY-SA 3.0
Names and complex mechanism is not important here. But I will describe it anyway. If you find this part boring, you can skip it. The basic idea (for eukaryotes) is that MSH and MLH class of proteins form protein complex (huge protein machines) that scans through DNA for any mismatches. If mismatches are found then, an protein called PCNA is recruited which further stabilises the mismatch region binding to the complex. Another enzyme exo1 which is an exonuclease that eats the damaged strand from near by nick is recruited. Both PCNA and exo1 helps in chewing up the damaged strand. Then another protein RPA is recruited which stabilizes the naked single stranded gap. In the meantime a DNA polymerase comes and fills in this gap (see Jun et al., 2006, for more details).
Shit happens - getting rid of chemically modified bases and lesions in DNA
But error made by your secretary is not the only time information in your documents can be damaged. What if someone spills a coffee on your documents, or burn it with Xray or UV light - well yeah, shit happens. In relation to DNA spilling of coffee could be equated to exposure to mutagen compounds like that from smoking, some food item or just random chemical event. But, how are you supposed to be able to read the instruction properly and make the right machines for your company from stained and burned documents? Well, neither can cell. So we need another set of employees that dedicate their life to keep an eye on these kind of damages and take a proper action.
Base excision repair - fixing the known modification of bases

Modified from Image by Lionel Allorge | CC BY-SA 3.0
For our cells these set of employees work in two departments - base excision repair (BER) and nucleotide excision repair (NER). The main difference lies in the repair strategy. The BER recognises the damaged bases that happen often and are well known to cells. These damages causes only slight distortion if DNA, and cells have specific proteins dealing with individual kind of damage. To put it in context, these damages are like fading of the ink on the paper, or someone ran a pen through the writing or made a tiny spot of ink. What you would do to take care of this in your office is - erase the problem with a whitener, then copy the info from the complementary copy of the document.
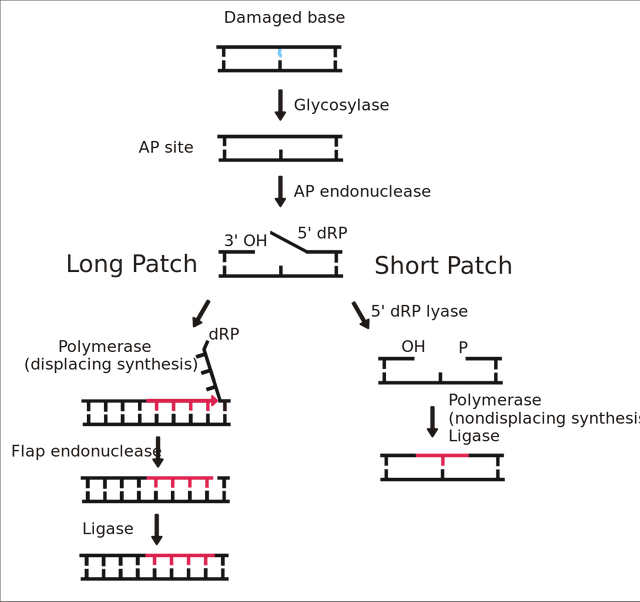
Image by Amazinglarry | Public domain
In the world of DNA, these kind of damages are related to spontaneous decay of DNA by oxidation, deamination, methylation, alkylation of the DNA bases, due to all the chemicals that are present in the cell, or due to some mutagen or carcinogenic shit you ate or smoked up. These reaction changes the bases by adding or remove some chemical groups to them, for instance oxidation of guanine forms 8-oxoguanine, or spontaneous deanimation of cytosine converts it to uracil. Since these mistakes occur via spontaneous decay of DNA, cells have evolved proteins that scans genome and specifically recognise these modification. For example, the uracil is recognised by uracil glycosylase and 8-oxoguanine by 8-oxoguanine glycosylase. Similarly, for all the modifications that occur there are specific glycosylases - 11 identified in mammals so far. Once they recognise the modified base they remove the base, leaving the sugar and phosphate backbone intact.
This is like having an employee who constantly checks the documents for faded ink, another one which check for a crossed letter, and a different one that finds a small stain hiding the text. When they find what they are looking for they erase that text from the page, but leave the page intact.
Then comes an endonuclease, which cuts this abasic site out of the strand. Usually known as AP endonuclease. This is like another employee who is called upon after text has been erased, they cut out the textless paper and calls for a sticker guy, who makes the sticker based on complementary document. Finally, you have a ligase, which seals the sticker with the paper. The sticker maker in case of DNA is DNA polymerase beta or epsilon depending in if its long patch repair or a short patch repair. (See Krokan and Bjørås, 2013 for details)
See this animation of BER-long patch
See this animation for BER short patch
Dealing with spilled coffee - repair of bulky lesions by nucleotide excision repair
On the other hand, NER deals with bulky lesions. Damages that cause big ass distortions in DNA. Such as those caused by exposure to UV light. For instance, if someone spilled a coffee on a file of your documents or say burned a part of paper in it. Then you need to excise the paper with burn or coffee stain, take a new paper, place it in the file and copy data from complementary page.
What you need in this case are employees which look for big stains in text. Or alternatively, someone reading the document can also inform the document verifier that there is a stain on the page. The verifier comes and recruit two men with scissors, who cut the page from opposite ends. The page is then excised out of the scroll. The new page is then copied from complementary copy and stuck in its place.
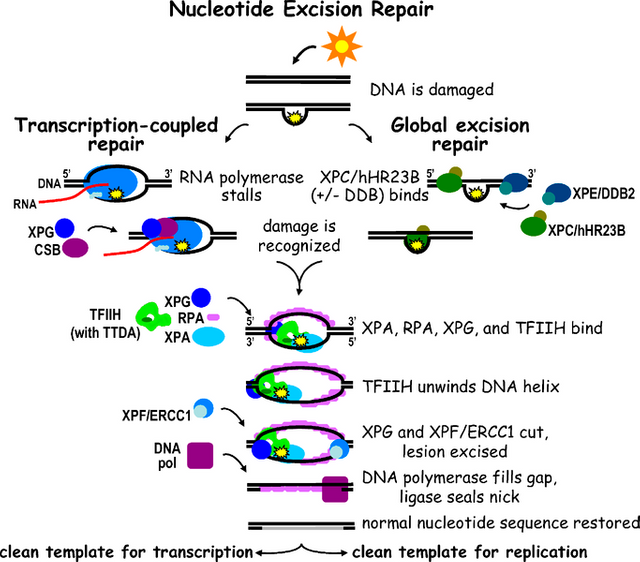
Image by Jill O. Fuss, Priscilla K. Cooper | CC BY 2.5.
The above description hints that there are two kinds of NER. One which happens by global scanning of genome and other which happens during reading (expressing) genes via RNA polymerase. Anyway, I should warn you that reading further is only important if you are interested in details. You can skip this and move to next section instead. The global scanner is a protein machine called XPC-RAD23B. It scans for any DNA distortions caused by bulky lesions. Alternatively, some lesions like thymine-dimers, made by UV light, are recognized by UV-Damage binding proteins 1 and 2. They remove the damaged dimer and instead put their amino acids there to create a wedge. This distortion is again picked up by XPC-RAD23B. This protein then recruits another complex protein machine called TFIIH consisting of about 10 different proteins. The job of this complex is to verify the damage and to pry open the two strands of DNA for other proteins to bind.
On the other hand the transcription coupled NER is initiated by a stalled RNA polymerase, which cannot move further on DNA because of these lesions. The stalled RNA pol eventually causes recruitment of TFIIH which takes further from there. In either case, once TFIIH is recruited it says goodbye to XPC-RAD23B. Instead now XPA, RPA, XPG, and ERCC1-XPF protein machines are recruited to the open strands of DNA. XPG and ERCC1-XPF acts as scissors and cuts the damaged strand from opposite ends excising it, and sending it to cellular oblivion, where it will be degraded eventually. In the meantime a DNA polymerase comes by and fills the gap. The newly synthesized strand is then finally sealed in place by a Ligase (See Schärer, 2013 for more details).
Dealing with torn off pages - Double stranded breaks.
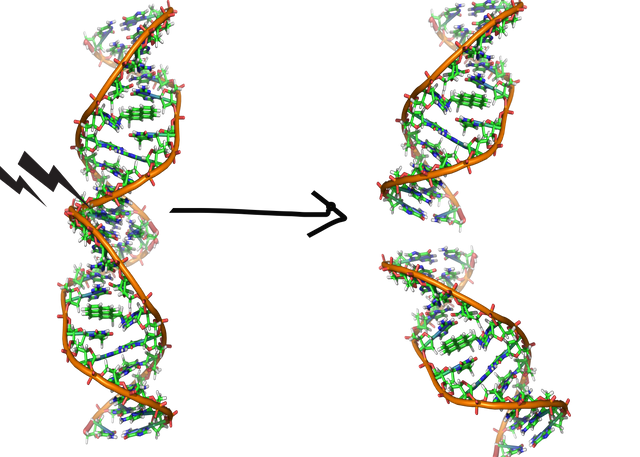
Image by Zephyris | CC BY-SA 3.0
Last, but not the least, it may so happen that someone might just tear off your documents - both the pages. Or in case of DNA break both the strands. This could be a real stressful situation, and you would want to put it back together as soon as possible. In context of DNA, this is a double stranded break, which can be caused either by spontaneous reaction or via some DNA damaging agent or via gamma irradiation. So, you need a set of protein employees dedicated to search for double stranded overhangs in the DNA.
Non-Homologus End Joining (NHEJ) - a quick fix
If it was your scroll of the documents that has been torn of what you would need first is - someone to see it. Then two employees which hold the two torn end together, bring them together and another one who seals it.
For a DNA double stranded breaks the free ends are first marked by ATM/ATR proteins. They phosphorylate the nearby histone H2AX (hitone are proteins on which DNA is wrapped) and causes binding of MRN complex. The free ends are then recognised by Ku70/80. They form a ring around broken ends of DNA. They then recruit two more machines namely - Artemis and DNA-PKcs. If the double strand break is blunt - that is it has no overhangs then DNA-PKcs will just phosphorylate 3 more machines - XLF, XRCC4 and Ligase4. The former two hold the ends together while aligase seals the break. However, in case there is a overhang, then you need another set of proteins to make the DNA end blunt. Artemis does that by chewing away the free hanging strand on one of the strand. While DNA pol lambda and mu does archives this by filling the end with complementary bases. Either way, once the ends are blunt you can seal them by process described above. Also, it is to be noted that a small overhang of 1-3 pair is created anyway at this site so that strands stick together better. (see Davis and Chen, 2013 and Shrivastav et al 2008 for more).
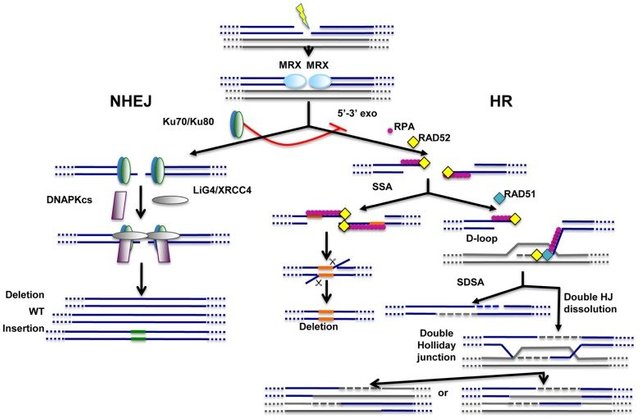
Image by Anabelle Decottignies, 2013 | CC BY 3.0
But, as you might have guessed that chewing away the ends and filling them before sealing might not always be perfect. Say, if there was an overhang that got chewed away but the other free end did not have the complementary overhang to recover the lost info, it might lead to error. Another scenario is where there might be more than two free ends of DNA swimming around in the nucleus. There is no way for these proteins to know which end goes with whom. They might just seal any two ends with correct polarity and boom, everything goes wrong from there. So while NHEJ may come handy, it has good chance of making errors. Which is why cells evolved and alternative pathway as well, so all the load is not always on this error prone pathway.
Homology Directed Repair (HR) - just more complicated stuff!
So what do you do in your office to circumvent this problem of sticking the wrong pages together? Well lucky you if once upon a time in history of offices they came up with an idea to have two copies of every thing. Yup that's all your diploid organisms who have two pair of chromosomes. So it's like having two identical scrolls or files of documents each having it's own pair complementary documents. So in case something happens to information on one scroll, you always have another to compensate. But you can also use this alternative scroll/file to fix the other one, can't you?
This is where homologous repair comes in. The idea is to identify the double stranded break. Find a homologous sequence and use it to fix the break. This is somewhat like two employees competing in who will fix the damage. So,if instead of Ku 70/80, an exonuclease, such as Exo1 or HO-nuclease is recruited it chews of one of the strands creating a long single stranded overhang. The overhang is then covered with RPA - the single strand stabilising protein. The Rad51 along with Rad52 and BRCA2 competes with RPA to bind the single stranded DNA. If successful, it initiates the search for homologous sequences and inserts the first strand into the homologous sequence, creating a D loop structure. The other end if possible can be brought to this site by another protein, such as Rad52. The invasion of other end then causes formation of a structure called holiday junction, which is then acted upon by other proteins, such as resolvase which eventually leads to repair of the damaged strands and sealing them together (see Jasin and Rothstein, 2013 for details)
Though one must remember that while homology directed repair maybe less error prone, it leads to another malady - the loss of hetrozygosity. If you don't know what that means, it simply means that it reduces the genetic diversity. So say if you had two different variant of sane gene that codes a protein to recognises viruses - you will be much better at dealing with different kind of virus infection than some who has exactly similar copy on both chromosomes. Homology repair kind of make both the copies alike.
Summary
In a nutshell, DNA faces wear and tear in all organisms. But all cells have evolved multiple strategies, which anticipate different kinds of damages and repair them. We saw that when mistakes happen during replication, protein machine belonging to mismatch repair system, senses the wrong topology of DNA caused by base-pair mismatch. They then recruit more machines to fix it. Then, there also a strategy to deal with damages that occur due to spontaneous decay of DNA bases, caused by endogenous or exogenous factors. Cells have machines specific for these kind of damages which belong to base excision repair system. This system works by removing the bases and then filling the gaps. On the other hand we saw yet another strategy for more bulky lesions. This one is less specific and takes care of anything that causes major distortion of DNA structure and topology. These machines implementing such strategy belong to nucleotide excision repair.
Then we saw the most toxic kind of DNA damage that exists - the double strand breaks. They are dealt with two competing repair strategies - non homologous end joining repair system, or to homology directed repair system.
Now it is to be noted that there are some other repair strategies that have not been discussed here. However this post does cover all major strategies found in all lifeforms we know.
Anyhow, the crux of the matter is that DNA damage that happens as we live our lives pose threat to our well being. When the damage happens in wrong place (an oncogene or tumor suppressor gene) it can cause cancer. Over the time the mutations and damages that accumulate in the cell also cause age related maladies. Which was topic of previous post. The idea was that the efficiency of these repair system goes down with age. Probably, because some of the proteins associated with DNA repair are downregulated - such as DNA pol beta, which helps in base excision repair. However, with caloric restriction the expression of these repair related genes can be pumped up back to decent levels. If that article was confusing to you before, you can give it a quick re-read once again.
About steemstem
But, before I go I would like to mention about the steemstem platform. Well, if you love reading and writing interesting science articles @steemstem is a community on steem that support authors and content creators in STEM field. If you wish to support steemstem do see the links below.
You can vote for steemstem witness here -Quick link for voting for the SteemSTEM Witness(@stem.witness)
Quick delegation links for @steemstem
50SP | 100SP | 500SP | 1000SP | 5000SP | 10000SP).
Delegating to @steemstem gives ROI of 65% of the curation rewards.
References
The quest for elixir of life - understanding caloric restriction (episode 1 - DNA damage and repair

Awesome post and blog! ;)
Thanks 🙂
Posted using Partiko Android
Hey @scienceblocks,
two cool scienceblocks articles within a few days!?! Marvelous!
I really appreciate that you try to explain the entire topic on an ordinary level. This enables people who are not thus far into this subject to imagine what's going on.
To be honest, I always forget all those mechanisms to fast. Therefore, I will resteem this and will find it much easier when I forgot a detail again ;-)
Have a nice weekend
Regards
Chapper
Yeah, I am hoping this is easier to understand for people not from the subject. And, thanks for the wonderful comment. Made my day. 🙂🤓
Posted using Partiko Android
Awesome overview of repair pathways. This stuff is closely aligned with my postdoctoral research and I'm quite impressed with how much info you packed in while keeping it accessible to everyone.
Wow, thanks. This comment means a lot coming from someone who did their postdoc in this topic. Made my day. :)
Posted using Partiko Android
Hi @scienceblocks!
Your post was upvoted by @steem-ua, new Steem dApp, using UserAuthority for algorithmic post curation!
Your UA account score is currently 3.426 which ranks you at #7260 across all Steem accounts.
Your rank has dropped 15 places in the last three days (old rank 7245).
In our last Algorithmic Curation Round, consisting of 177 contributions, your post is ranked at #116.
Evaluation of your UA score:
Feel free to join our @steem-ua Discord server
This post has been voted on by the SteemSTEM curation team and voting trail. It is elligible for support from @curie and @utopian-io.
If you appreciate the work we are doing, then consider supporting our witness stem.witness. Additional witness support to the curie witness and utopian-io witness would be appreciated as well.
For additional information please join us on the SteemSTEM discord and to get to know the rest of the community!
Thanks for having added @steemstem as a beneficiary to your post. This granted you a stronger support from SteemSTEM.
Thanks for having used the steemstem.io app. You got a stronger support!
Hi @scienceblocks!
Your post was upvoted by Utopian.io in cooperation with @steemstem - supporting knowledge, innovation and technological advancement on the Steem Blockchain.
Contribute to Open Source with utopian.io
Learn how to contribute on our website and join the new open source economy.
Want to chat? Join the Utopian Community on Discord https://discord.gg/h52nFrV
Amazing how are the molecules that carry our information; I read an article of @rebe.torres12 where Japanese people built a DNA of 8 blocks, demonstrates that the exchange of information in molecules is common and possible.